原子力显微镜
AFM配件
应用
联系我们
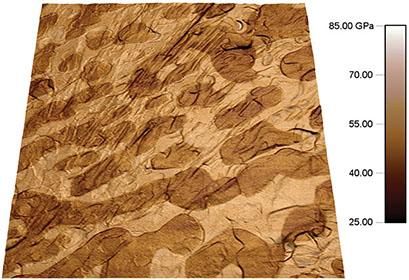
纳米级的机械性能是许多应用中考量的一个关键因素,而原子力显微镜则是能够测量这些性能的工具之一。利用Asylum Research 的NanomechPro™工具包,你可以测量任何材料(从细胞到陶瓷)的纳米级机械特性。这些技术能够准确地评估各种纳米级机械行为,包括弹性、粘性、附着力和硬度。NanomechPro™工具包的多种技术可以为不同的应用提供更大的灵活性,并通过结果比对实现更深入的理解。此外,利用NanomechPro™工具包的模式,还可以对更多特性进行更快速的测量。NanomechPro™工具包支持Cypher™ 和MFP-3D™ 系列原子力显微镜的功能。
咨询AFM领域的专家"Probing the swelling-dependent mechanical and properties of polyacrylamide hydrogels through -based dynamic nanoindentation," Y. Lai and Y. Hu, Soft 14, 2619 (2018). https://doi.org/10.1039/c7sm02351k
"Controlling the mechanoelasticity of model biomembranes with room- ionic liquids," C> Rotella, P. Kumari, B. J. Rodriguez, S. P. Jarvis, and A. Benedetto, Biophys. Rev. 10, 751 (2018). https://doi.org/10.1007/s12551-018-0424-5
"Tendon exhibits complex poroelastic behavior at the nanoscale as revealed by - -based rheology," B. K. Connizzo and A. J. Grodzinsky, J. Biomech. 54, 11 (2017). https://doi.org/10.1016/j.jbiomech.2017.01.029
"Polymer nanomechanics: Separating the size from the substrate in nanoindentation," L. Li, L. M. Encarnacao, and K. A. Brown, Appl. Phys. Lett. 110, 043105 (2017). https://doi.org/10.1063/1.4975057
"Mechanical properties of highly porous super liquid‐repellent surfaces," M. Paven, R. Fuchs, T. Yakabe, D.,Vollmer, M. Kappl, A. N. Itakura, and H.-J. Butt, Adv. Funct. Mater. 26, 4914 (2016). https://doi.org/10.1002/adfm.201600627
"Practical loss tangent imaging with amplitude-modulated atomic force microscopy," R. Proksch, M. Kocun, D. Hurley, M. Viani, A. Labuda, W. Meinhold, and J. Bemis, J. Appl. Phys. 119, 134901 (2016). https://doi.org/10.1063/1.4944879
"Fast, quantitative nanomechanical measurements using AM-FM Viscoelastic Mapping mode," D. Hurley, M. Kocun, I. Revenko, B. Ohler, and R. Proksch, Microscopy and Analysis 29, 9 (2015). Download Here
"Contact resonance atomic force microscopy imaging in air and water using photothermal excitation," M. Kocun, A. Labuda, A. Gannepalli, and R. Proksch, Rev. Sci. Instrum. 86, 083706 (2015). https://doi.org/10.1063/1.4928105
"Predictive modelling-based and experiments for synthesis and spinning of bioinspired silk fibres," S. Lin, S. Ryu, O. Tokareva, G. Gronau, M. M. Jacobsen, W. Huang, D. J. Rizzo, D. Li, C. Staii, N. M. Pugno, J. Y. Wong, D. L. Kaplan, and M. J. Buehler, Nat. Comm. 6, 6892 (2015). http://doi.org/10.1038/ncomms7892
"Nano-rheology of hydrogels using direct drive force modulation atomic force microscopy," P. C. Nalam, N. N. Gosvami, M. A. Caporizzo, R. J. Composto, and R. W. Carpick, Soft 11, 8165 (2015). https://doi.org/10.1039/c5sm01143d
"Fast nanomechanical of soft ," E. T. Herruzo, A. P. Perrino, and R. Garcia, Nat. Commun. 5, 3126 (2014). https://doi.org/10.1038/ncomms4126
" intrinsic mechanical flexibility of mouse prion nanofibrils revealed by measurements of axial and radial 's moduli," G. Lamour, C. K. Yip, H. Li, and J. Gsponer, ACS Nano 8, 3851 (2014). https://doi.org/10.1021/nn5007013
"Quantifying cell-to-cell variation in power-law rheology," P. Cai, Y. Mizutani, M. Tsuchiya, J. M. Maloney, B. Fabry, K. J. V. Vliet, and T. Okajima, Biophys. J. 105, 1093 (2013). https://doi.org/10.1016/j.bpj.2013.07.035
"Nanomechanical mapping of soft by bimodal force microscopy," R. Garcia and R. Proksch, Eur. Polym. J. 49, 1897 (2013). https://doi.org/10.1016/j.eurpolymj.2013.03.037
"Loss tangent imaging: and simulations of repulsive-mode tapping atomic force microscopy," R. Proksch and D. G. Yablon, Appl. Phys. Lett. 100, 073106 (2012). https://doi.org/10.1063/1.3675836
"Mapping nanoscale elasticity and dissipation using dual contact resonance ," A. Gannepalli, D. G. Yablon, A. H. Tsou, and R. Proksch, 22, 355705 (2011). https://doi.org/10.1088/0957-4484/22/35/355705
"Mapping nanomechanical properties of live cells using multi-harmonic atomic force microscopy," A. Raman, S. Trigueros, A. Cartagena, A. P. Z. Stevenson, M. Susilo, E. Nauman, and S. A. Contera, Nat. Nanotechnol. 6, 809 (2011). https://doi.org/10.1038/nnano.2011.186
"Viscoelastic property mapping with contact resonance force microscopy," J. P. Killgore, D. G. Yablon, A. Tsou, A. Gannepalli, P. Yuya, J. Turner, R. Proksch, and D. C. Hurley, Langmuir 27, 13983 (2011). https://doi.org/10.1021/la203434w
"Mechanical properties of face-centered cubic supercrystals of nanocrystals," E. Tam, P. Podsiadlo, E. Shevchenko, D. F. Ogletree, M.-P. Delplancke-Ogletree, and P. D. Ashby, Nano Lett. 10, 2363 (2010). https://doi.org/10.1021/nl1001313
"Tuning the elastic modulus of hydrated collagen fibrils," C. A. Grant, D. J. Brockwell, S. E. Radford, and N. H. Thomson, Biophys. J. 97, 2985 (2009). https://doi.org/10.1016/j.bpj.2009.09.010
"Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength," B. C. Isenberg, P. A. DiMilla, M. Walker, S. Kim, and J. Y. Wong, Biophys. J. 97, 1313 (2009). https://doi.org/10.1016/j.bpj.2009.06.021
" viscoelasticity of individual gram-negative bacterial cells measured using atomic force microscopy," V. Vadillo-Rodriguez, T. J. Beveridge, and J. R. Dutcher, J. Bacteriol. 190, 4225-4232 (2008). https://doi.org/10.1128/jb.00132-08
"A -layer model for viscoelastic, stress-relaxation testing of cells using atomic force microscopy: Do cell properties reflect metastatic potential?" E. M. Darling, S. Zauscher, J. A. Block, and F. Guilak, Biophys. J. 92, 1784 (2007). https://doi.org/10.1529/biophysj.106.083097
"Packing density and structural heterogeneity of insulin amyloid fibrils measured by nanoindentation," S. Guo, and B. B. Akhremitchev, Biomacromolecules 7, 1630 (2006). https://doi.org/10.1021/bm0600724
"Multifrequency, repulsive-mode amplitude-modulated atomic force microscopy," R. Proksch, Appl. Phys. Lett. 89, 113121 (2006). https://doi.org/10.1063/1.2345593
 公安机关备案号31010402003473
公安机关备案号31010402003473
